Genes and Genetics of Tuberculosis-Juniper publishers
JUNIPER PUBLISHERS-OPEN ACCESS INTERNATIONAL JOURNAL OF PULMONARY & RESPIRATORY SCIENCES
Abstract
Tuberculosis affects human life globally for long
time. About one third of the world’s population is infected with the
causal pathogen Mycobacterium tuberculosis but without presenting any
clinical symptoms. The difference in clinical outcome of infection
suggests that host genetic makeup is responsible for such variability.
Attempts have been made to identify the underlying genes. In case of
Mendelian susceptibility to mycobacterial disease (MSMD) a rare disease
with immune-deficiency, mutations were identified in genes that impair
IFN γ signaling pathway. Linkage studies have identified several loci
but exact gene was never pinpointed. Candidate gene association studies
carried out in different populations, identified several risk alleles.
But findings of all these studies were hardly replicated in another
population. Findings are also not justifiable in some cases because of
limited sample size. GWAS also identified several susceptible locus,
many of which were replicated in another population where as many were
not. Gene expression analysis also adds onto identification of gene
implicated in infection and thus enhances knowledge on genes playing
significant role in mycobacterium infection. However all these studies
show that not a single gene but many genes are orchestrated together in
determining the fate of infection. More research is necessary to find
out such genes, their interaction with other members and complicated
network formed.
Abbreviations: FEV1: Forced Expiratory Volume in the First Second; FVC: Forced Vital Capacity;, PEF: Peak Expiratory Flow; MVV: Maximum Voluntary Ventilation; FEF25-75%: Forced Expiratory Flow Rate Over 25-75% Part of FVC; DSE: Diaphragm Strengthening Exercise; sEMG: Surface Electromyogram; RMS: Root Mean Square
Tuberculosis (TB) is a deadly disease afflicting
human kind from long time. Pre historic evidence found in excavation
[1], mummies [2] etc. suggest that tuberculosis was present in ancient
days. The disease is caused by infection with Mycobacterium tuberculosis
which is transmitted from an infected person in form of aerosol
droplets. WHO estimates 10.4 million new cases in 2015 [3]. There is an
estimate of 1.4 million deaths due to TB in the year 2015. The outcome
of infection is manifold. Only a minor group of people develop active
tuberculosis upon exposure to Mycobacterium tuberculosis. A handful of
individuals are able to clear the infection, whereas majority of
infected individuals harbor the infection in latent condition.
In latent condition Mycobacterium within macrophages
encloses itself in cellular aggregates formed by different kind of
immune cells. Such compact cellular aggregates are called granuloma [4].
About one third of the world population belongs to latent infected
group. Only 10% of them may express the disease in one’s life time by
reactivation of the latent pathogen depending on the immune status of
the host. All these observations, lead to the obvious question why such
differences exist and what determines such differences? Before
discoveryof mycobacterium bacilli by Robert Koch, it was thought that
tuberculosis has a prominent hereditary component as many members in the
same family were affected. However, after Koch’s discovery the thought
was that Mycobacterium tuberculosis is sole responsible for the disease
and elimination of the bacteria will prevent the disease. But it was
gradually realized that bacilli alone is not sufficient for an
individual to express the disease.
At present, it is unequivocally proved that not only
the pathogen but host factors have major contribution in successful
establishment of infection. Twin studies and animal models suggest that
host factor play considerable role in predisposing an individual to such
infections. Concordance of tuberculosis is higher among monozygotic
twins than dizygotic twins [5,6]. Animals infected with M.tb can result
into susceptible and resistant group depending on the genetic background
of the animal. All these evidences suggest that host susceptibility is
determined by the genetic makeup of an individual which controls immune
response. Genetic locus controlling such susceptibility to infection can
be identified by screening and comparing infected individuals in a
family or community to non-infected individuals [7], by comparing
syntenic locus identifiedin animals [8] already demonstrated to have
role in infection or
genes which has functional implication in immunity [9].
With the aim to identify genes or genetic variants playing
role in susceptibility to tuberculosis, initially linkage study
and later on case control association studies were undertaken
by several groups. Linkage studies are family based, where
affected individuals sharing similar phenotypes as well as
unaffected members are screened for genetic markers across
all chromosomes. Commonality of markers in different regions
of chromosomes of the affected members, are compared to
unaffected members to locate a genetic loci which is significantly
linked and co-segregate with the phenotype. The genomic locus
identified may harbor the causal gene that contributes to such
altered disease phenotype. The case control approach is another
approach where instead of family members, unrelated ‘case’
(active disease) and ‘control’ (without disease) are enrolled for
screening genetic markers. It is then tested whether the genetic
variants are associated with the disease trait. Selectively few
candidate genes can be tested for the purpose or the test may be
extended to the whole genome level.
Mendelian Susceptibility to Mycobacterium Disease (MSMD)
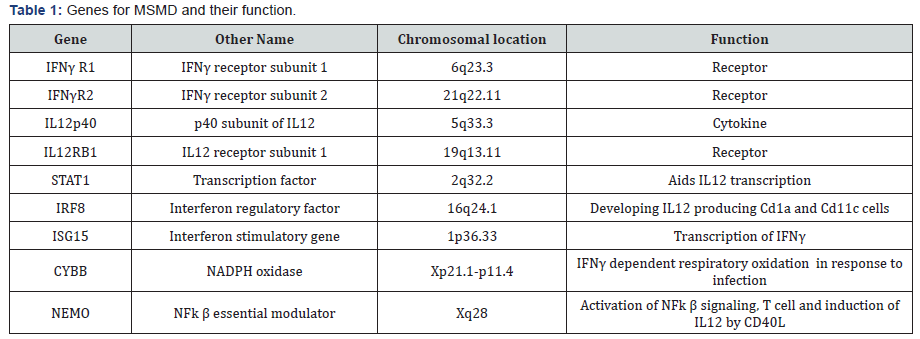
A rare form of tuberculosis known as Mendelian
Susceptibility to Mycobacterium Disease (MSMD) is a class of
disease where children are immune compromised, displaying
severe symptoms of tuberculosis even when infected with weak
strain of mycobacterium like BCG or natural atypical strains
[10]. The penetrance of the disease is highly variable. Linkage
studies with the affected family members led to identification of
the region on chromosome 6 harboring the gene IFNγ receptor.
Sequencing of IFNγ receptor 1 gene identified mutations leading
to premature termination [11]. Later on mutations were detected
in IFN-γR2 gene also [12]. The two subunits combine to form
IFN-γ receptor, which binds to its ligand IFNγ and transduce the
signal to downstream effector molecule. Eventually many more
mutations were identified in several other genes which include
autosomal genes like IRF8, IL12B, IL12RB1, STAT1, ISG15
and X-linked NEMO, CYBB [13] (Table 1). The common thread
between these genes is that they all are involved in the circuit
of IL12 induced IFNγ activation pathway [14]. These mutations
lead to recessive or dominant form of disease, with complete or
partial loss of function. Mutations in all above mentioned genes
explain 50% of the cases with remaining 50% cases still unknown
for mutations. All these mutations cause inactivation of IFNγ or
impaired signaling leading to inborn error in immunity. This
suggest that IFNγ mediated immunity is central to mycobacterial
infection.
Linkage studies
Shaw and his colleagues studied several families in Brazil
and identified a TB linked locus presenting weak linkage to
CXCR2 gene (P= 0.039) which is tightly linked to SLC11A1
(NRAMP1) gene [15]. This region encompasses SLC11A1
and TNF gene cluster even though neither of them presented
any evidence of linkage independently. SLC11A1 was well
characterized and known for its variants to be associated
with tuberculosis. Hypothesizing that this gene may be a good
candidate gene for susceptibility to tuberculosis a linkage study
was performed in a large Aboriginal Canadian family. Evidence
of linkage was found in SLC11A1 region (2q35) (LOD=3.8) but
no mutations were reported [16]. Another outcome of this
study was that no significant linkage to HLA region was found
which is otherwise thought to have important role in infection.
Gambian and South African sib pair analysis identified seven
loci, two of which (15q11-q13 LOD 2.00, Xq26 LOD 1.77) were
replicated in another independent set. However, all of these
linkages were weak [17,18]. Further evidence of linkage was
obtained at the locus 8q12-13 (LOD>3) in a study performed on
96 Morroccan families [19]. Stein et al reported linkage to 7p22
locus among Ugandan people, which harbors IL6 gene nearby [20]. They also reported additional two loci 2q21-24 and 5p13-
5q22 associated with phenotype of non reactivity to tuberculin
skin test. Evidence of age specific variation was obtained in a
study led by Mahasirimongkol et al. [21]. They reported linkage
in two regions 17p13.3-13.1, 20p13-12.3 in patients from
Thailand, when patients were stratified on the basis of age of
onset (<25 yr). Several groups later undertook fine mapping of
the regions identified by linkage analysis by typing more dense
markers in the region i.e. single nucleotide polymorphisms
(SNP). Fine mapping of 17q11-17 revealed, presence of many
genes like NOS2A, CCL2/MCP-1, CCL3/MIP-1a, CCL4/MIP-1b,
CCL5/RANTES, CCR7, STAT3 and STAT5A/5B in the region [22].
Screening of this region showed evidence of linkage with LOD
score of 2.48 (p 0.0004). Similarly, fine mapping of region 5q31
which spans Th2 cytokine gene cluster revealed association
of haplotypes with tuberculosis [23]. Failure to replicate the
identified loci in other populations, has dampened the findings
of linkage study. This also suggests that not a single gene but
multiple genes determine susceptibility to tuberculosis.
Candidate gene association studies
More than three hundred reports describe association of
tuberculosis with DNA variants in more than hundred candidate
genes. Candidates are chosen based on their role in immunity.
Highly reported and well studied few genes are SLC11A1, VDR,
TLRs, HLA class II molecules, IFNg, IL10, TNFa [9]. The most
successful and convincing study is with SLC11A1 gene which has
been replicated in several countries. Studies on mouse model
identified and mapped a locus on chromosome1 controlling
infection towards mycobacterium, salmonella, and leishmania.
The identified gene was called Bcg and later renamed as Nramp1
(natural resistance associated macrophage protein, also known
as Solute Carrier Family 11a member1 SLC11A1). The human
homologue of Nramp1 was identified and mapped to chromosome
2q35. NRAMP1 is a metal transporter localized in late endosome
of macrophages and recruited to phagosome when phagocytosis
occurs. Evidence of significant linkage to NRAMP1 or SLC11A1
was demonstrated in a large indigenous Canadian family [16].
The four variants of SLC11A1 gene INT4, D543, 3’UTR, 5’GT as
risk allele for TB have been studied in several populations across
the globe [24]. Effect of each of the variant is highly variable
among different population [25]. Some variants of SLC11A1 not
only represent high degree of susceptibility to tuberculosis, but
also severe form of it [25,26,27]. Meta analysis suggests that
variants in SLC11A1 are significantly associated with Asian and
Africans with PTB but not among people of European origin
[28]. The genetic variants of SLC11A1 are strongly associated
(OR 1.75(CI 1.10-2.77), p=0.01) with tuberculosis susceptibility
among children [29].
Vitamin D level inversely correlates with severity of TB [30].
It is well established that Vitamin D plays role in defense against
mycobacterium by inducing antimicrobial peptide cathelicidine,
an inducer of autophagy in macropahage and boosting adaptive
immunity. Vitamin D also modulates differentiation and growth of different immune cells. All these cells express vitamin D
receptor through which Vitamin D acts. High doses of Vitamin
D along with normal course of drugs are used for tuberculosis
treatment. Four well known DNA variants in Vitamin D receptor
(VDR) gene are studied among different population. They are
designated as Fok1 (rs10735810), BsmI (rs154410), Apa I
(rs7975232), TaqI (rs731236) depending on the ability of the
restriction enzymes to cut at the specific locations. The FokI
site has a C/T polymorphisms which determine the amount of
VDR produced and contributes to risk for tuberculosis (OR =
1.507, 95%CI = 1.192-1.906, P = 0.001). Meta-analysis suggests
that the roles of other polymorphisms are not significant with
development of pulmonary tuberculosis [31] among East Asians.
These polymorphic sites are located in the 3’UTR and may have a
role in VDR mRNA stability. The results with VDR polymorphisms
are also inconsistent among different population. Few studies
including the study by Lombard et al did not reveal any
association of tuberculosis with VDR polymorphisms, but the
F-b-A-T haplotype was observed as a protective factor for TB
in South Africa [32]. Other haplotypes f-T-B and f-T-B as risk for
tuberculosis were reported in Iranian population [33].
A study on Indians from northern part reported association
of HLA-DR2 to susceptibility to tuberculosis [34]. Although
no association or linkage was detected in population from
south India [35]. Association of HLA DQ alleles are reported
in Cambodia [36]. Among Iranian patients HLA-DRB1*07 and
HLA-DQA1*0101(OR 2.7, 95%CI 1.19-6.13, P= 0.025 and OR
2.66, 95%CI 1.15-6.44, P = 0.04, respectively) appeared to be
the predisposing alleles and HLA-DQA1*0301 and 0501 the
protective alleles (OR 0.254, 95%CI 0.075-0.865, P = 0.033
and OR 0.53, 95%CI 0.3-0.95, P = 0.045, respectively) [37].
Associations of different alleles to susceptibility and protection
have been reported in South Africa, Greece, Poland, and China.
Analysis of (HLA)-DRB1 and -DQB1 gene polymorphisms
among Koreans suggest that DRB1*0803 (OR = 1.97, p = 0.012)
and DQB1*0601 (OR = 2.07, p = 0.005, p(c) > 0.05) alleles are
associated with progression of tuberculosis to severe form and
development of drug resistance [38]. A study led by Salei M
attempted to correlate HLA class I polymorphism of the host with
the strain of M.tb infected [39]. Further they have shown that
presence of HLA-B27 allele protect an individual from another
episode (OR=0.21 p=0.006) of disease. More recently a metaanalysis
including 31 study suggests that the HLA-DRB1*04, *09,
*10, *15, and *16 gene polymorphisms [*04 (OR 1.22, 95% CI
1.00-1.48, P = 0.048), *09 (OR 1.50, 95% CI 1.08-2.08, P = 0.016),
*10 (OR 1.23, 95% CI 1.01-1.49, P = 0.035), *15 (OR 1.40, 95%
CI 1.14-1.73, P = 0.001), and *16 (OR 1.33, 95% CI 1.08-1.63, P
= 0.007)] may be associated with risk of TB, particularly among
the East Asian. But the HLA-DRB1*11 gene polymorphism *11
(OR 0.72, 95% CI 0.53-0.99, P = 0.044), may have protective role.
No significant association between the HLA-DRB1*01, *03, *07,
*08, *12, *13, and *14 gene polymorphisms and TB risk was
found [40].
Toll like receptor (TLR) play important role in activation
of innate immunity against mycobacterial infection. Pathogen
associated molecular patterns (PAMP) are recognized by TLRs.
These receptors are present on cell surface or intracellularly in
cytoplasm or on endosomal membranes. TLR2 and TLR4 form
heterodimer with TLR1 or TLR6 and recognize mycobacterial
components. Polymorphisms in TLR genes are extensively
studied to test association with tuberculosis susceptibility in
different ethnicities, but results are contradictory. rs 4833095
in TLR1 gene is associated with resistance to tuberculosis. Metaanalysis
suggest that heterozygous individuals with AG genotypes
are protected than GG (AG vs. GG: OR=0.77,95% CI=0.65-0.95,
p=0.0031) [41]. On meta-analysis rs5743708 turned out to be
non significant, even though individual studies report A allele
as a risk allele for Hispanic and Asian population. Analysis of
another SNP in TLR2 gene (rs3804100) demonstrated that CC
genotype is risk for developing tuberculosis. Variants in TLR4
(rs4986791), TLR6 (rs5743810), TLR9 (rs352139) turn out to
be risk or protective when studied individually, but overall do
not pose any strong effect on risk for TB development.
This study explored the effect of 12 weeks of strengthening
exercise of diaphragm in asthmatic children. It revealed a
statistical significant higher values of FEV1, FEV1/FVC, PEF
and MVV and also a significant increase in diaphragmatic
thickness and excursion in asthmatic patients after diaphragm
strengthening exercise. Many authors studied the effect of non
respiratory exercise on pulmonary function parameters in
asthmatic children [19, 21,26-29,] Their results revealed marked
improvement in pulmonary functions after diaphragmatic
training. Girodo [29] studied the effect of 16-week program of
diaphragmatic strengthening exercise for asthmatic patients.
They found a significant reduction in medication use and in the
intensity of asthmatic symptoms. A follow up at two months
found that many patients have returned to earlier medication
levels with marked impairment in their PEF. McCool [30]
postulated that weight-bearing maneuvers may be used to
strengthen the diaphragm and expiratory muscles. However
they found that although strength training leads to myofiber
hypertrophy, it does not result in mitochondrial proliferation, so
weight lifting increases diaphragm structure and pressures with
less effect on contractility and excursion.
The candidate association studies performed on different
populations are highly heterogeneous in nature. In many studies
it is reflected that age should be given importance and age turned
out to be an important factor. The variable results of association
may be due to genetic heterogeneity, clinical heterogeneity and
different LD pattern in different population and limited sample
size in each study.
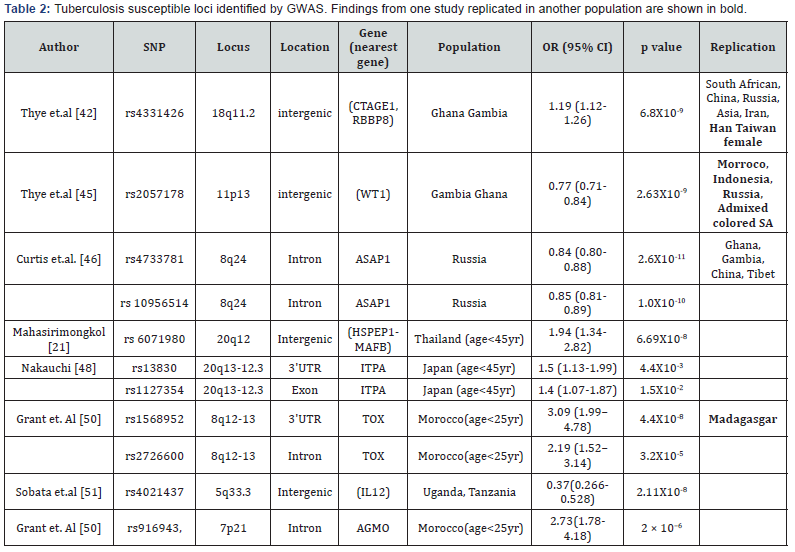
Genome wide association studies (GWAS)
The aim of Genome wide association study is to identify
disease associated DNA variants in a genome wide manner in
a large number of samples. Cases with disease and controls
without disease are compared in GWAS with appropriate
precautionary measurements. The first GWAS for tuberculosis
done on African population from Ghana and Gambia, identified
an intergenic SNP rs 4331426 (OR 1.19 (1.12-1.26), p=6.8X10-9)
on the chromosomal region 18q11.2 [42]. However the biological
implication of this SNPS was not known as it is located in the
gene desert region. The same tested in Chinese population was
significant but with opposite effect (p= 0.011, OR 0.62 (0.44-
0.87) i.e. protective as reported by Wang et.al. [43]. This locus
also failed to replicate in South African Colored population [44].
Another study on Ghana and Gambian population identified
another non coding SNP rs 2057178 (OR 0.77 (0.71-0.84),
p=2.63X 10-9) which was associated with resistance to TB [45].
The nearest genes WT1 and RCN1 were located 45Kb and 500
Kb respectively both of which apparently have no connection
with infection. However association of this SNP was validated
in Russian (p = 2.0 × 10−2, OR 0.91, (0.82-0.99), Indonesian (p=
9.9 × 10−2, OR 0.84, (0.68-1.03), African Colored population (p=
2.71X 10-6, OR 0.62 (0.5-0.75) [44] also. Since then few more
studies across different countries and population have identified
some more locus.
A GWAS performed on Russian identified several significant
SNPs at the locus 8q24. The most significant variant rs 4733781
(p=2.6X10-11 OR 0.84 (0.8-0.88), is located in an intron of
ASAP1 gene. It was demonstrated that this variant can alter
ASAP1 expression in dendritic cell affecting its migration
[46]. Significant association of rs 4733781 also hold true for
African population from Ghana and Gambia, but not in Western
Chinese Han and Tibetan population [47]. However the Russian
population did not show any association with the SNP at the
locus 18q11 previously reported in African population. Another
interesting observation was that the significant SNPs identified
in African population apparently failed to replicate in Thai
and Japanese population. An earlier study on Thai population
presented evidence of linkage on 20q12. But only when the
patients were stratified based on their age (cut off of 45yr), the
young tuberculosis patients presented significant association
with SNP rs 6071980 (p=6.69X10-8 OR 1.94 (1.34–2.82)) on
20q12 [21]. The nearest genes HSPEP1-MAFB are potential
candidates for TB susceptibility. Recently, deep sequencing of the
region 20q13-12.3 identified rs13830 and rs1127354 in ITPA
gene showing association with young (age < 45 yr) TB patients
[48] in Japan. The region 5q31 harbor a gene cluster of Th2
cytokine and showed evidence of linkage for tuberculosis earlier.
Fine mapping of this region among Thai trio families identified
DNA variants in three genes SLC22A4, SLC22A5 and KIF3A of
nominal significance. However haplotype constructed with three
markers from these genes remain significant even after multiple
testing corrections [23]. This implies that multiple DNA variants
play role in tuberculosis. A separate study on Indonesians
identified nine independent locus near genes JAG1, DYNLRB2,
EBF1, TMEFF2, CCL17, HAUS6, PENK and TXNDC4. Findings of
this study were validated in another Indonesian group as well as
among Russian [49] independently or in combination but none
of them attained genome wide significance. Previously reported
susceptible loci 8q12-13 in a family based discovery study from
Morocco was further densely mapped by genotyping SNPS
located in the region [19]. Two SNPs rs1568952 and rs2726600
located in introns of TOX gene were significantly associated
with tuberculosis (combined p = 1.1 × 10-5 and 9.2 × 10-5). The
association was even stronger in patients with age less than 25
yr. TOX is required for the development of the CD4 T lineage.
Results were replicated in Madagascar nuclear families with
early onset of TB [50].
A comparative study was performed in a cohort consisting of
people from Uganda and Tanzania, consisting of HIV coinfected
TB patients and only HIV infected individual who do not develop
tuberculosis infection in spite of close exposure to TB patients
[51]. A SNP rs4021437 at 5q33.3 was significantly associated
with TB infected individuals (OR 0.37, p =2.11X10-8) in a HIV
positive background. This SNP is located near IL12 gene and
embedded in H3K27Ac his tone mark possibly indicating its
role in regulation. This again strengthens the fact that IL12 has
important role in TB infection. Summary of GWAS done are given
in Table 2.
Host response and activation of genes
Host response to infection is reflected in its transcriptional
signature. Altered gene expression also provide clue for
identifying host genes implicated in infectious disease. In case of
tuberculosis host gene expression profile has been studied using
whole blood or PBMC or different immune cells [52,53,54]. Many
of these studies have concluded similar type of genes altered in
tuberculosis infection and can discriminate active disease or
latency or even from any other type of infection. Altered genes
are majorly immune regulator, cytokines or receptors or involved
in inflammation or apoptosis triggered by pathogen infection
[55]. A study involving patients from UK and Africa suggested
presence of neutrophil driven interferon signature with active
TB cases, which is absent in latent and healthy individuals [53].
This signature was validated in samples from different countries
and different assay platforms. This study also demonstrated
that there was significant change in transcriptomics after
two months of treatment. Two other studies in Africa also
demonstrated decline in certain transcripts after administration
of drugs [56,57]. Complement genes within this list suggest
complement mediated decrease in bacterial load [57]. Different
mycobacterium strain can evoke differential immune response.
Change in gene expression was monitored in lung epithelial cells
after infecting with different strains of mycobacterium [58].
Strain specific signature was visible with overlapping signature
as well. Strain specific signature and activation of functional
pathways were also observed for two strains. Strain specific
signatures are of immense importance to identify strain specific
biomarkers and immunotherapy.
It is clear that host response to any infection is a multistep
process. It is a complex interaction between host and pathogen.
In order to understand the biology of infection one needs to
dissect the complex interaction between the host genes which
are activated to protect the host, whereas the pathogen genes
counteract the host defense mechanism. Development of modern
genomic tools has enabled us to understand the molecular
events. HIV infection has aided the spread of tuberculosis and
so is diabetes. Even though we have knowledge on association of
numerous variants and their role in tuberculosis but many more
are yet to be discovered. Also in many cases it is not clear how
the hits of GWAS contribute to susceptibility. More studies are
required in future for better understanding.
To know more about Open Access International
Journal of Pulmonary & Respiratory Sciences please click on: https://juniperpublishers.com/ijoprs/index.php
To know more about Open access Journals
Publishers please click on : Juniper Publishers


Comments
Post a Comment